The MiCell( C++ | Java ) interface is the most important interface used to define the cell topology and thus the topology of unstructured meshes. Non linear cells can only be defined via the MiCell( C++ | Java ) interface. Independent of the cell dimension (1D, 2D, 3D) the following methods must be implemented for non linear cells (as for linear cells):
getNumEdges: Must return the number of edges of the cell. This number must be independent of the order of the cell. For instance a triangle cell must always return 3 even if it is a quadratic triangle, a cubic triangle etc.
getNumNodes: Must return the number of nodes of the cell. This number depends on the order of the cell. For instance a linear triangle cell must return 3, a quadratic triangle cell must return 6 etc.
getWeight(pcoord, weight) : Must return the value of the shape functions (weight) for the given iso parametric coordinates (pcoord). The method is not pure virtual, but the default implementation throws an exception. Thus the method must be overridden for non linear cells.
getWeight(geometry, coord, weight) : Alternative method returning the shape functions for the given point (coord) in world coordinates. An implementation of this method should first compute the iso-parametric coordinate of coord (by using the given mesh geometry), then call the first form of getWeight.
getIsoParametricCoord (nodeId): must return the iso parametric coordinates of the given nodeId. The iso parametric coordinates belong to an interval that must be consistent with the definition of the shape function. Usually the interval is [-1,+1] or [0,1] but it is not mandatory.
The MiCell( C++ | Java ) interface has 3 derived interfaces to get more specific properties for 1D cells, 2D cells and 3D cells.
getNumEdges(): this inherited method should always return 1 for 1D cells.
size_t getSubEdgeNodesIndex(vector<size_t>& edgeNodeIds): This method must return the number of sub edges linking all the nodes of the non linear cell. For instance a quadratic 1D cell must return 2, and a cubic 1D cell must return 3. It returns 0 by default (meaning definition of a linear cell) and must be overridden for non linear 1D cells. The edgeNodeIds array must be filled by a list of pairs of local node identifiers (one pair per sub edge). Local identifiers are identifiers defined in the range 0 to getNumNodes()-1.
For instance, edgeNodeIds should be filled with the 4 values {0,2,2,1} for the following quadratic 1D cell:
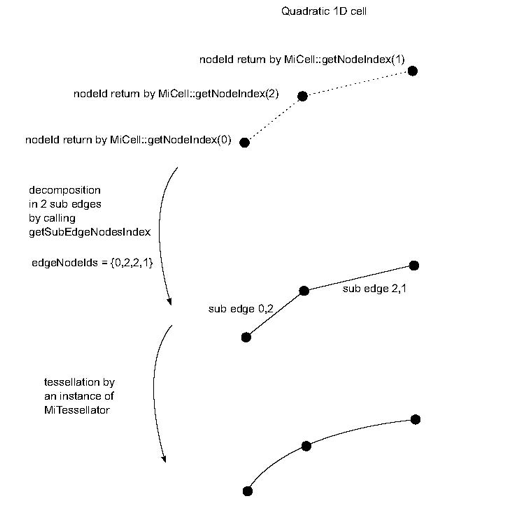
As shown in the previous figure, the instance of MiTessellator( C++ ) tessellates each sub edge given by getSubEdgeNodesIndex by calling MiTessellator( C++ )::tessellateLineCell()
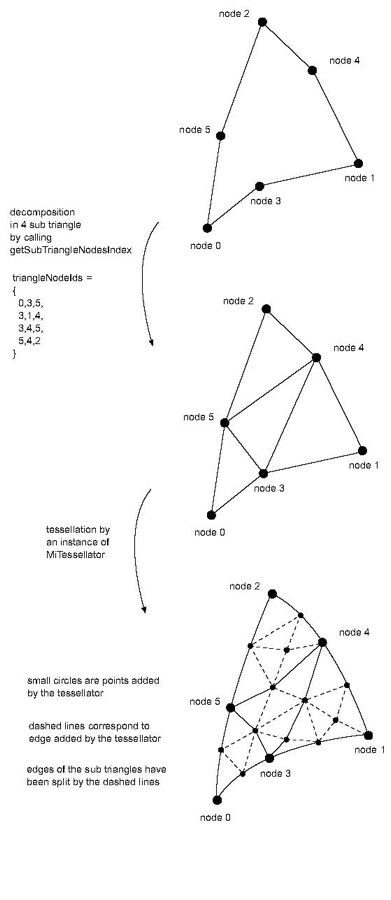
size_t getSubTriangleNodesIndex (vector<size_t>& triangleNodeIds): This method must give a decomposition of the surface cell in sub triangles. The set of sub triangles must fill all the area of the non linear surface cell. The method must return the number of sub triangles linking all the nodes of the non linear cell. For instance a quadratic triangle cell must return 4. It returns 0 by default (meaning definition of a linear cell) and must be overridden for non linear 2D cells. The triangleNodeIds array must be filled by a list of triplets of local node identifiers (one triplet per sub triangle). Local identifiers are identifiers defined in the range 0 to getNumNodes()-1.
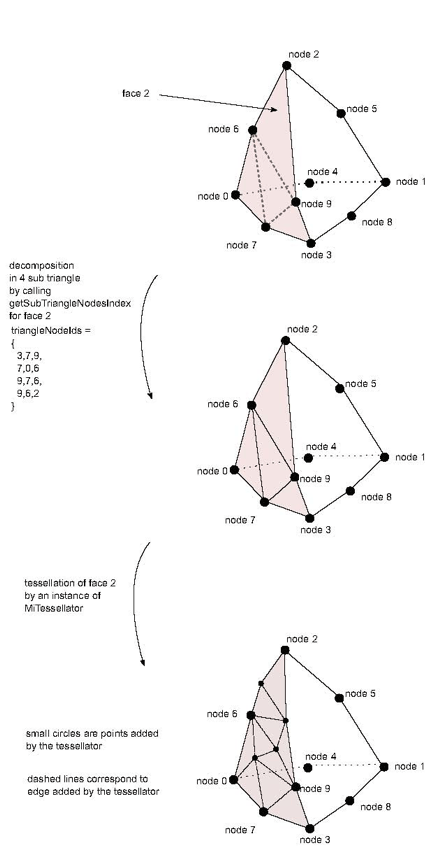
size_t getSubTriangleNodesIndex (faceId, vector<size_t>& triangleNodeIds): A method similar to MiSurfaceCell( C++ | Java )::getSubTriangleNodesIndex but for one face of the volume cell. This method must return the number of sub triangles linking all the nodes of the given faceId of this non linear cell. For instance calling getSubTriangleNodesIndex for one of the triangle faces of a quadratic tetrahedron cell must return 4. It returns 0 by default (meaning definition of a linear cell) and must be overridden for non linear 3D cells. The triangleNodeIds array must be filled by a list of triplets of local node identifiers (one triplet per sub triangle). Local identifiers are defined in the range 0 to getNumNodes()-1.
size_t getSubTetrahedronNodesIndex (vector<size_t>& tetrahedronNodeIds): This method must give the decomposition of the non linear volume cell into sub tetrahedrons. The set of sub tetrahedrons must fill all the volume of the non linear volume cell. It is called for instance by the isosurface extractor or the plane slice extractor. It is not used by the skin extractor.
This method must return the number of sub tetrahedrons linking all the nodes of this non linear volume cell. It returns 0 by default (meaning definition of a linear cell) and must be overridden for non linear 3D cells. The tetrahedronNodeIds array must be filled by a list of quadruplets of local node identifiers (one quadruplet per sub tetrahedron). Local identifiers are defined in the range 0 to getNumNodes()-1.
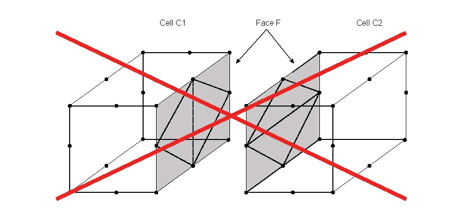
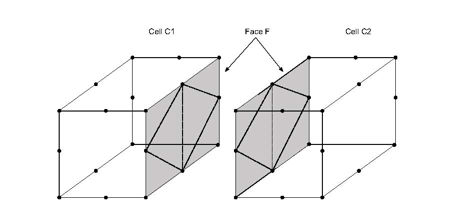
Attention: The triangle faces of the returned sub tetrahedrons must match the triangle decomposition returned by MiVolumeCell( C++ | Java )::getSubTriangleNodesIndex. The triangle faces of sub tetrahedrons of one cell C must also match the triangle faces of sub tetrahedrons of an adjacent cell to C. For instance, in the following figures the decomposition of the face F by the sub tetrahedra of the cell C1 must match the decomposition of the face F by the sub tetrahedra of the adjacent cell C2.
For instance the decomposition of a quadratic hexahedron of 27 nodes should return 48 sub tetrahedrons.
See the section called “Properties of shape functions” for a more detailed information about the definition of iso parametric coordinates, the weight (aka the shape functions) and the properties they must respect.
MeshViz provides several utility classes that help to implement the most common finite element cells. As with defining shape functions, defining the sub tetrahedron decomposition is not always trivial, these classes give an easy to use implementation of these tasks. Depending on the cell dimension, these utility classes give a static implementation of the most useful methods of respectively MiLineCell( C++ | Java ), MiSurfaceCell( C++ | Java ) or MiVolumeCell( C++ | Java ) interface.
These utility classes are:
MxTriangleCellExtract
MxTriangle6CellExtract
MxQuadrangleCellExtract
MxQuadrangle8CellExtract
MxQuadrangle9CellExtract
MxTetrahedronCellExtract
MxWedgeCellExtract
MxWedge18CellExtract
MxPyramidCellExtract
MxHexahedronCellExtract
MxHexahedron20CellExtract
MxHexahedron27CellExtract
The utility classes with a number in the class name correspond to the implementation of quadratic cell (one additional node per edge), and this number is the number of nodes in the cell. The utility classes without a number in the class name correspond to the implementation of linear cell.
These classes have the following common properties:
They define only static methods and don’t have any non static data.
They implement getIsoParametricCoord by returning the iso parametric coordinates for each node of the cell. These iso parametric coordinates are defined in the interval [0-1]. Notice that this interval is a convention used only by these classes Mx<Shape>CellExtract. But MeshViz can work with any other convention for the interval used by iso parametric coordinates, (provided the shape functions follow the 3 properties defined previously)
They implement getIsoParametricCoord by computing the iso parametric coordinates of a given point in the world coordinates space.
They implement getWeight(…) defining the shape functions from the iso parametric coordinates in the same interval [0-1]
They implement isPointInsideCell returning true if a point given in world coordinate is inside the cell.
Furthermore, the linear volume cell extract class implements the getIsosurfTopology which gives the marching cube decomposition of the cell.
The following code gives an example of using the MxHexahedron20CellExtract( C++ ) class to implement a quadratic hexahedron cell of 20 nodes:
C++
class Hexa20Cell : public MiVolumeCell
{
public:
...
virtual void getWeight(const MbVec3d &ipcoord,
std::vector<double>& weight) const
{
return MxHexahedron20CellExtract::getWeight(ipcoord,weight);
}
virtual void getWeight(const MiGeometryI& meshGeometry,
const MbVec3d &coord,
std::vector<double>& weight) const
{
return MxHexahedron20CellExtract::getWeight(meshGeometry,this,coord,weight);
}
virtual size_t getSubTetrahedronNodesIndex(std::vector<size_t>& tetrahedronNodeIds) const
{
return MxHexahedron20CellExtract::getSubTetrahedronNodesIndex(tetrahedronNodeIds);
}
...
}
See the demonstration program QuadraticHexa20 to see the full implementation of this example class Hexa20Cell.
The following screen shots show miscellaneous results of using the Mx<Shape>CellExtract classes.
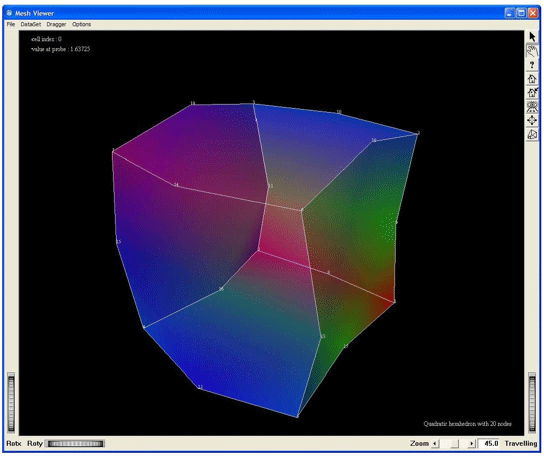
Visualization of a quadratic hexahedron cell of 20 nodes, by using the MxHexahedron20CellExtract. This screen shot shows the cell without any tessellation.
Figure 2.34. Demo QuadraticHexa20:
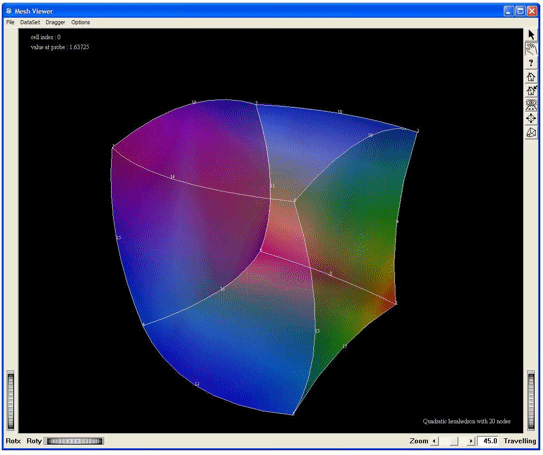
Visualization of a quadratic hexahedron cell of 20 nodes, by using the MxHexahedron20CellExtract. This screen shot shows the cell with a geometric tessellation.
Figure 2.35. Demo QuadraticHexa20:
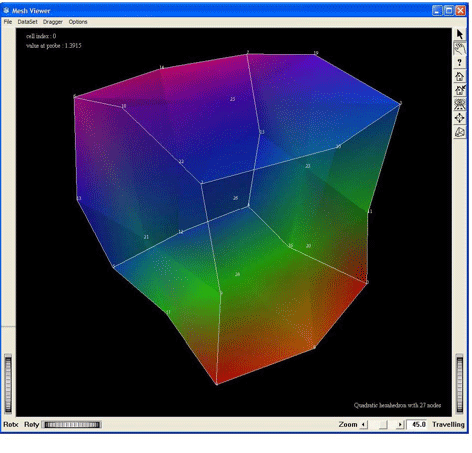
Visualization of a quadratic hexahedron cell of 27 nodes, by using the MxHexahedron27CellExtract. This screen shot shows the cell without any tessellation
Figure 2.36. Demo QuadraticHexa27:
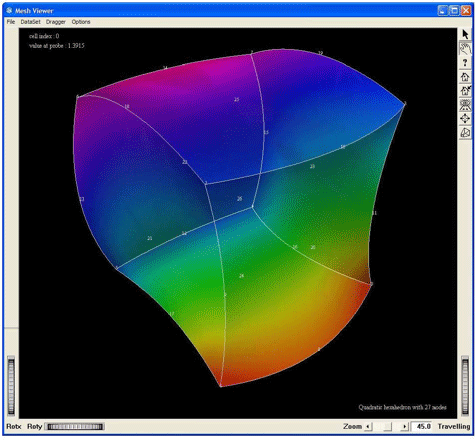
Visualization of a quadratic hexahedron cell of 27 nodes, by using the MxHexahedron27CellExtract. This screen shot shows the cell with a geometric tessellation.
Figure 2.37. Demo QuadraticHexa27:
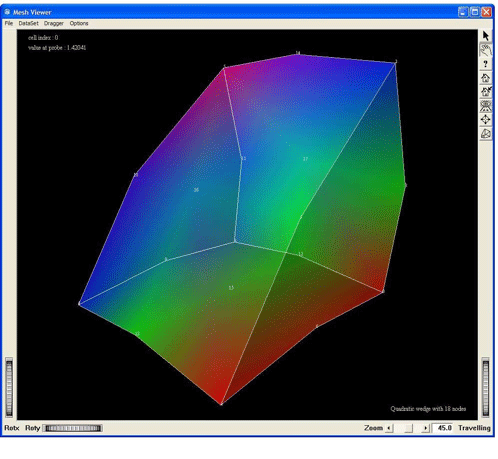
Visualization of a quadratic wedge cell of 18 nodes, by using the MxWedge18CellExtract. This screen shot shows the cell without any tessellation
Figure 2.38. Demo QuadraticWedge18:
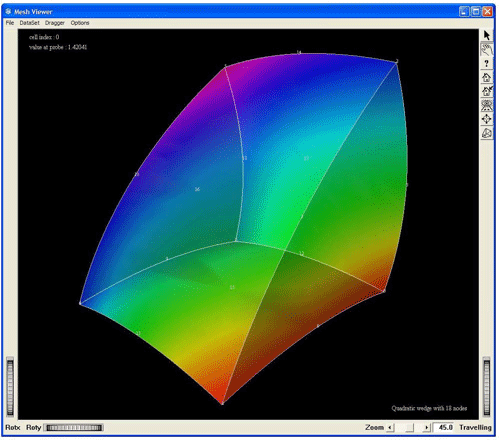
Visualization of a quadratic wedge cell of 18 nodes, by using the MxWedge18CellExtract. This screen shot shows the cell with a geometric tessellation
Figure 2.39. Demo QuadraticWedge18: