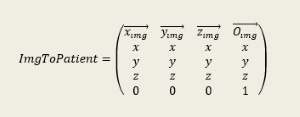DICOM is a widely used format for storing medical image data (CT, MRI, etc), defined by the National Electrical Manufacturers Association (NEMA) (http://medical.nema.org). The SoVRDicomFileReader( C++ | Java | .NET ) class can load a volume from a single DICOM file or from a list of DICOM files (stack of images). Loading a volume from a single DICOM file is the same as loading any other format supported by VolumeViz. Loading a volume from a list of DICOM files can be done using a list file or by specifying a list of filenames. Using a list file is the same as loading a stack of images using the SoVRRasterStackReader( C++ | Java | .NET ) except the list file should not contain any header. Each line in the list file may be a full path containing directory names or a simple file name. Simple file names are assumed to be in the same directory as the list file. The application can also specify a list of file paths using the setFilenameList() method. This is useful, for example, if the application user is allowed to select a list of files in a file selection dialog.
Unlike a raster stack, the position of each slice in the volume is determined by the location value in its file header and not by the order of the file name in the list. Also unlike a raster stack consisting of (for example) JPEG or PNG images, a DICOM file in the list may contain more than one slice of the volume. The reader handles this automatically. The first file in the list is considered the “reference” for the volume and all slices must be compatible with this one. Specifically this means that subsequent files must have the same width, height, data type and so on. The volume is oriented in 3D space in a natural way for medical images. The slice width is along the X axis, the slice height is along the Y axis and slice location values increase along the Z axis.
Note when using a list file: If the file extension is not “.dc3”, “.dic” or “.dicom” VolumeViz will not automatically select the DICOM volume reader. You can either give the list file one of the DICOM file extensions or force VolumeViz to use the DICOM reader by creating an instance of SoVRDicomFileReader( C++ | Java | .NET ) and calling the setReader() method as described in the “Non-standard extension” sub-section of Section 1.2.3, “Loading from a file”.
The volume reader will automatically get the correct volume dimensions, data type, extent (voxel size/spacing) and number of significant bits from the DICOM file header. The reader will also apply the data adjustment (if any) specified by the RescaleSlope and RescaleIntercept tags in the DICOM file header, i.e.: actualValue = slope * storedValue + intercept. As part of this process the reader will automatically convert unsigned data to the corresponding signed data type if necessary (in other words if the rescale calculation produces negative values). The application can also explicitly specify the volume data type. This allows, for example, converting float data values to more compact integer values.
![[Important]](../../images/important.jpg) | |
The DICOM reader only uses the rescale slope and intercept values from the first file in the list. It does not currently handle the (less common) case where each file contains different rescale values. |
The SoVRDicomData( C++ | Java | .NET ) class allows the application to access DICOM specific data in the file header(s). One way to do this is to query the volume reader from the SoVolumeData( C++ | Java | .NET ) then query the SoVRDicomData( C++ | Java | .NET ) object from the reader. However the application can also create an instance of SoVRDicomData( C++ | Java | .NET ) explicitly and then open DICOM files directly using the readDicomHeader() method. SoVRDicomData( C++ | Java | .NET ) provides methods such as getSliceSpacing() to query some of the commonly used values. However it also provides the getDicomInfo() method which allows the application to query any DICOM tag (if present in the file) by its hexadecimal group and tag number.
C++
SoVRDicomFileReader* pReader = (SoVRDicomFileReader*)pVolData->getReader(); const SoVRDicomData &dicomData = pReader->getDicomData(); float rescaleSlope = dicomData.getDefaultSlope(); SbString str = dicomData.getDicomInfo( 0x28, 0x1053 ); if (! str.isEmpty()) { rescaleSlope = str.toFloat(); }
.NET
SoVRDicomFileReader Reader = (SoVRDicomFileReader)VolData.GetReader(); SoVRDicomData dicomData = Reader.GetDicomData(); float rescaleSlope = dicomData.GetSlope(); string str = dicomData.GetDicomInfo(0x28, 0x1053); if (str.Length > 0) { rescaleSlope = float.Parse(str); }
Java
SoVRDicomFileReader reader = (SoVRDicomFileReader)volData.getReader(); SoVRDicomData dicomData = reader.getDicomData(); float rescaleSlope = dicomData.getSlope(); String str = dicomData.getDicomInfo((short)0x28, (short)0x1053); if (str.length() > 0) { rescaleSlope = Float.valueOf(str); }
Displaying a DICOM as a RAW image on a screen is easy. Locating properly a DICOM in a 3D scene may be much more tricky. This part is not dedicated to OpenInventor but helps to understand how DICOM are located in spaces.
If you are a 3D computer graphics or if you are using OpenInventor, you may already be familiar with the concept of 3D vector space. You probably heard about World Space, Camera Space, Projection Space, Object Space, etc. We will now introduce some new spaces specific to DICOM world.
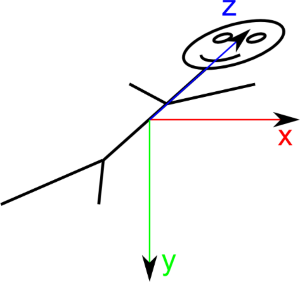
DICOM defines a term: "Reference Coordinates System" or RCS, also called “Patient Space”. Patient Space is defined as follow for BIPED specimen like humans:
X direction is from Right to Left of patient
Y direction is from Antero to Posterior, or Front to Back of patient
Z direction is from Foot to Head of patient
X, Y and Z are normalized. Origin of patient space is at (0, 0, 0). This form a Direct trihedron folowing the the Right-Hand rule:
Another mnemonic way to name this space is “RL-AP-FH” for Right/Left-Antero/Posterior-Foot/Head.
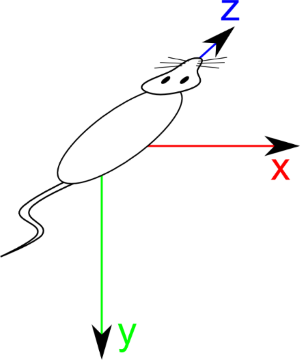
For QUADRUPED specimen like mouses, space is defined as follow:
X direction is from Left to Right of patient
Y direction is from Dorsal to ventral
Z direction is from Foot to Head of patient
This actually correspond to natural way to introduce specimen inside an MRI or a scanner. Type of specimen is defined by DICOM tag Anatomical Orientation Type (0010,2210). If this tag is absent, specimen is considered BIPED. Starting from now, type of specimen has no more importance as everything will be defined from “Patient Space” or “RCS”.
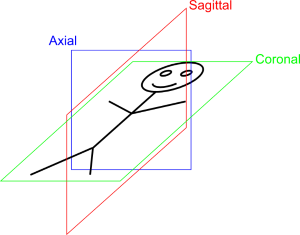
From this patient space, we can derive the 3 main cut Sagittal normal to X axis, Coronal normal to Y axis and Transverse (aka Axial) normal to Z axis:
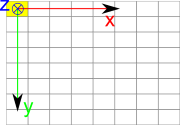
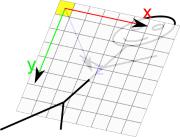
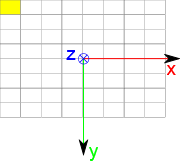
DICOM define an ImageSpace relative to image. This space is defined as follow:
X is along image line
Y is along image coloumn
Z is cross product of X and Y, so that based is orthogonal direct
Origin is in center of top left pixel
Vectors have a unit length of 1mm
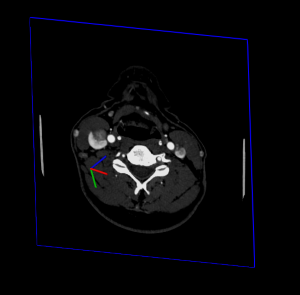
On the image below, yellow pixel is pixel which is supposed to be displayed in Top Left on the screen.
Now the difficulty consist in properly locate image in patient space. Unfortunatly, image plane are not always aligned with Sagital, Coronal or Axial plane but can be oblique:
So, permuting X, Y and Z axes or applying just a translation is not enough. One must also take in account image rotation given by the tag Image Orientation (0020, 0037). This tag gives the coordinates of X and Y image axes in Patient Space. So the proper transformation to locate image in patient space is given by the following matrix:
Where:
(O_img) : Image Position (Patient) (0020, 0032)
(X_img) : row (X) direction cosine of Image Orientation (Patient) (0020,0037). This vector is normalized with a length of 1mm
(Y_img) : col (Y) direction cosine of Image Orientation (Patient) (0020,0037). This vector is normalized with a length of 1mm
(Z_img) : Cross product between (x_img) and (y_img). This vector is normalized with a length of 1mm
More details can be found here
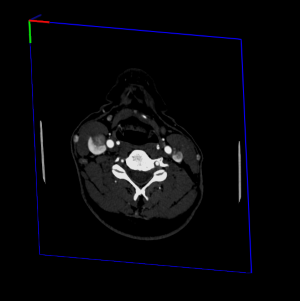
By default VolumeViz will locate DICOM images so that center of image will be in (0, 0, 0):
This may be useful in case of simple image viewer but may not be adapted for application using several 3D object and locate them in space. To properly locate image in image space or in patient space, you should call the MedicalHelper( C++ | Java | .NET )::dicomAdjustVolume(volumeData) method. This method exist in two versions:
C++
dicomAdjustVolume( SoVolumeData* volume, SbBool useImagePosition = TRUE );
This version works only with axis aligned volume. When specifying useImegPosition = FALSE, volume extent will be adjusted so that origin will be in center of top left pixel as defined in DICOM Image Space. When specifying useImagePosition = TRUE, image will be located so that center of top left pixel will be located in ImagePositionPatient (0020, 0032). This correspond to locating image in Patient Space, in case of axis aligned acquisition.
C++
dicomAdjustVolume( SoVolumeData* volume, SoMatrixTransform* imgToPatient );
This version is more generic and handle non axis aligned acquisitions. This method will adjust volumeData extent so that image will be properly located in image space, ie origin in center of top left pixel, and will compute the ImgToPatient matrix in an SoMatrixTransform( C++ | Java | .NET ). This node can be added in scenegraph just before SoVolumeData( C++ | Java | .NET ) to properly locate volume in patient space.
In following examples the small trihedron is located in scenegraph world origin
Example of image position without putting SoMatrixTransform( C++ | Java | .NET ):
And same example with SoMatrixTransform( C++ | Java | .NET )