 Defines an abstract cell of a mesh.
More...
Defines an abstract cell of a mesh.
More...
#include <MeshVizXLM/mesh/cell/MiCell.h>
 Inheritance diagram for MiCell:
Inheritance diagram for MiCell:Public Member Functions | |
| virtual | ~MiCell () |
| Virtual destructor. | |
| virtual size_t | getNumNodes () const =0 |
| Gets the number of nodes. | |
| virtual size_t | getNodeIndex (size_t node) const =0 |
| Gets the index (in the mesh) of a node. | |
| virtual double | getRelativeSize (const MiGeometryI *meshGeometry) const |
| Gets the relative size of the cell. | |
| virtual void | getWeight (const MbVec3d &ipcoord, std::vector< double > &weight) const |
| Gets the weights of a point defined by its iso parametric coordinates. | |
| virtual MbVec3d | getIsoParametricCoord (size_t nodeIndex) const |
| Gets the iso parametric coordinates of a cell's node. | |
| virtual MbVec3d | getCenter (const MiGeometryI &geometry) const |
| Gets the center of the cell. | |
| virtual bool | isPointInsideCell (const MiGeometryI &meshGeometry, const MbVec3d &point, std::vector< double > &weights) const |
| Checks if a point is inside or outside a cell. | |
Protected Member Functions | |
| virtual std::ostream & | toStream (std::ostream &s) const |
| Performs an output operation on a stream which is returned afterward. | |
Friends | |
| std::ostream & | operator<< (std::ostream &s, const MiCell &cell) |
| Outputs the mesh to a stream using the toStream protected method implemented in all derived classes. | |
 Defines an abstract cell of a mesh.
Defines an abstract cell of a mesh.
A cell is a basic volume, surface or edge element that connects adjacent nodes of a mesh. The MiCell interface is necessary to define the topology of unstructured 1D, 2D or 3D meshes.
MiCell is the base class that defines topological characteristics of a cell (characteristics not depending on the cell dimension).
These methods must be overridden by the appplication as they are pure virtual. The characteristics that depend on the cell dimension are defined in the derived interfaces MiLineCell for 1D cells, MiSurfaceCell for 2D cells, and MiVolumeCell for 3D cells.
MiCell also defines additional methods with a default implementation. A default implementation is provided in 2 cases:
The implementation of all the methods of this class must be thread-safe because it might be called concurrently by multiple threads. If the implementation can't be thread-safe, MeshViz XLM must be forced to use only 1 thread. It can be done by calling MiMeshViz::init(1) or MoMeshViz::init(1) or setting the parallel argument to false for each extractor instantiation. For instance by calling MiSkinExtractUnstructured::getNewInstance(&mesh, false);
|
inlinevirtual |
|
inlinevirtual |
Gets the center of the cell.
This method is not pure virtual because it is not used by all extraction classes. The default implementation provided computes the average of the coordinates of each cell's node.
| [in] | geometry | The geometry of the mesh. Used to retrieve the node coordinates of this cell. |
|
inlinevirtual |
Gets the iso parametric coordinates of a cell's node.
Only classes defining non linear cells must implement this method.
The interval value of parametric coordinates are usually [0,1] or [-1,+1]. If the application uses interval [0,1], the parametric coordinates returned by this method for linear cell must be either 0 or 1. For quadratic cell, the parametric coordinates must be either 0, 0.5 or 1. (0.5 for intermediate nodes).
See the following image as example of parametric coordinates values for linear and quadratic quadrangle.
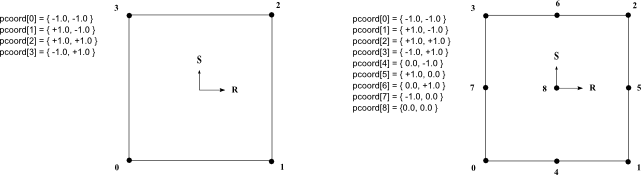
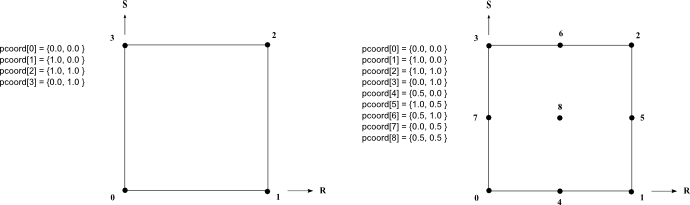
This method is not pure virtual because it is not used by all extraction classes. However using an extraction class that uses this method will generate an exception. An application does not need to override this method if no such extraction class is used.
| [in] | nodeIndex | The index of the node. |
|
pure virtual |
Gets the index (in the mesh) of a node.
| [in] | node | Node number (where: 0 <= node < getNumNodes()). |
|
pure virtual |
Gets the number of nodes.
This number must include the additional nodes of non linear cells. For instance getNumNodes() for a quadratic triangle cell should return 6.
|
inlinevirtual |
Gets the relative size of the cell.
This method is used only by the MoMeshCellShape to display node names if using the relative offset feature and the streamline extraction to adapt the step integration to the size of the cell. getRelativeSize() should return a value depending on the size of the cell even if the default implementation returns 1. For instance, getRelativeSize() can return the length of the longest edge of this cell, or the length of its diagonal.
If getRelativeSize() returns a constant value for any cell, the stream line extraction uses a constant number of step integration anywhere in the mesh. Returning a constant implies that all the cells are supposed to have the same size. getRelativeSize() must not return 0.
| [in] | meshGeometry | the geometry of the mesh. Used to retrieve the node coordinates of this cell. |
|
inlinevirtual |
Gets the weights of a point defined by its iso parametric coordinates.
Only classes defining non linear cells must implement this method.
A weight value (aka shape function) must be given for each node of the cell. The weight must respect the following rules:
See chapter "Properties of shape functions" in the User's Guide for more details. This method is not pure virtual because it is not used by all extraction classes. However using an extraction class that uses this method will generate an exception. An application does not need to override this method if no such extraction class is used.
| [in] | ipcoord | iso parametric coordinates of the points where the weights must be evaluated. |
| [out] | weight | Must contain at least as many elements as the number of nodes in this cell. |
weight[i] = wifor each i with 0 <= i < getNumNodes()
|
inlinevirtual |
Checks if a point is inside or outside a cell.
This method is not pure virtual because it is not used by all extraction classes. However using an extraction class that uses this method (i.e. streamline) will generate an exception. An application does not need to override this method if no such extraction class is used.
| [in] | meshGeometry | The geometry of the mesh. Used to retrieve the node coordinates of this cell. |
| [in] | point | The point to be checked. |
| [out] | weights | Must contain at least as many elements as the number of nodes in this cell (see getWeight()). |
weight[i] = wifor each i with 0 <= i < getNumNodes()
|
inlineprotectedvirtual |
Performs an output operation on a stream which is returned afterward.
This method is protected since only called by the output operator <<. This method is not pure virtual and output by default the geometry and topology of a mesh. Whenever a different output is needed, this method can be implemented in the subclass to get a different output format. No std::ostream& operator << needs to be defined in any subclass. This allows you to benefit from the virtual inheritance when using the output operator on an abstract class.
Reimplemented in MiVolumeCell.
|
friend |