SoMarkerBasedWatershedProcessing Class Reference
[Region Growing]
 SoMarkerBasedWatershedProcessing engine performs a fast determination of the watershed lines.
More...
SoMarkerBasedWatershedProcessing engine performs a fast determination of the watershed lines.
More...
#include <ImageViz/Engines/ImageSegmentation/RegionGrowing/SoMarkerBasedWatershedProcessing.h>
Public Types | |
| enum | OutputMode { SEPARATED_BASINS = 0, WATERSHED_RIDGES = 1 } |
| enum | PrecisionMode { REPEATABLE = 0, FAST = 1 } |
Public Member Functions | |
| SoMarkerBasedWatershedProcessing () | |
Public Attributes | |
| SoSFEnum | computeMode |
| SoSFEnum | neighborhood3d |
| SoSFEnum | outputMode |
| SoSFImageDataAdapter | inGrayImage |
| SoSFImageDataAdapter | inMarkerImage |
| SoSFEnum | precisionMode |
| SoImageVizEngineOutput < SoSFImageDataAdapter, SoImageDataAdapter * > | outObjectImage |
Detailed Description
 SoMarkerBasedWatershedProcessing engine performs a fast determination of the watershed lines.
SoMarkerBasedWatershedProcessing engine performs a fast determination of the watershed lines.
For an introduction, see Watershed principle.
SoMarkerBasedWatershedProcessing has 2 configurations. See outputMode field.
This engine performs a fast determination of the watershed lines from specified markers in a label image, followed by a labelling of the complementary set. It can be viewed as the zones of influence of the markers.
Either the separated basins are output in a label image, the crest lines being part of the complementary (not a label), or the crest lines are output in a binary image.
Note: Because two bits are used to code the pixels during the algorithm, input gray levels image should range from 0 to 16383. If the range is greater, a normalization is applied to reduce the dynamic of this image.
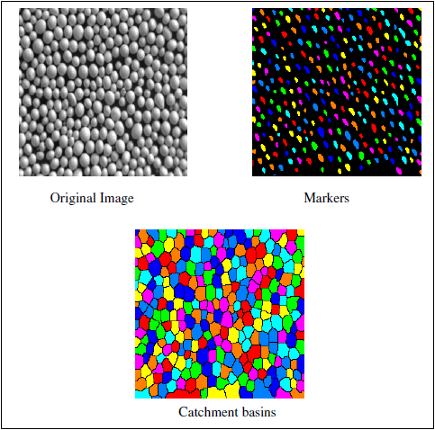
Example of SoMarkerBasedWatershedProcessing engine with SEPARATED_BASINS mode
Notice: This engine requires to preliminarily load the whole input data set into memory to be computed. As a consequence the input data set will be automatically loaded before the execution of this engine, ignoring the maximum memory parameter of the SbImageDataAdapterHelper::getAppropriateAdapter function or SoFileDataAdapter class If the input data set cannot fit in memory, this engine will fail during its computation. |
SEE ALSO
SoAddObjectToLabelProcessing, SoGrayscaleResconstructionProcessing
FILE FORMAT/DEFAULT
- MarkerBasedWatershedProcessing {
| computeMode | MODE_AUTO |
| neighborhood3d | CONNECTIVITY_26 |
| outputMode | WATERSHED_RIDGES |
| inGrayImage | NULL |
| inMarkerImage | NULL |
| precisionMode | REPEATABLE |
Library references: catchbasin fastwatershed
- See related examples:
Member Enumeration Documentation
Constructor & Destructor Documentation
| SoMarkerBasedWatershedProcessing::SoMarkerBasedWatershedProcessing | ( | ) |
Constructor.
Member Data Documentation
Select the compute Mode (2D or 3D or AUTO) Use enum ComputeMode.
Default is MODE_AUTO
The input grayscale image.
Default value is NULL. Supported types include: grayscale image.
The input marker (label) image.
Default value is NULL. Supported types include: label color image.
In 3D configuration (see computeMode), the neighborhood connectivity defines the connectivity considered for processing adjacent voxels.
Use enum Neighborhood3d. Default is CONNECTIVITY_26.
| SoImageVizEngineOutput<SoSFImageDataAdapter,SoImageDataAdapter*> SoMarkerBasedWatershedProcessing::outObjectImage |
The output image.
Default value is NULL. Supported types include: binary image.
Select the type of output mode.
Use enum OutputMode. Default is WATERSHED_RIDGES
Select the mode of algorithm precision for watershed.
Use enum PrecisionMode. Default is REPEATABLE
The documentation for this class was generated from the following file:
- ImageViz/Engines/ImageSegmentation/RegionGrowing/SoMarkerBasedWatershedProcessing.h